OpenFOAM
Enabling
Search through the available modules to see what OpenFOAM ones exist
$ module avail openfoam
This only shows versions that are compatible with other loaded
modules. All versions can be found with the spider command along
with directions on what else needs to be loaded to load them
$ module spider openfoam
$ module spider openfoam/11
Note that there two releases of OpenFOAM. One is openfoam.com and uses vYYMM (year and month) version numbering. The other is openfoam.org and uses X and X.Y version numbering. With the exception of a some specific features, the two are largely similar.
Load the openfoam module (not specifying a version uses the default). Note
that spider may shom additional modules that you need to load first.
$ module load gcc
$ module load openfoam
Can use the show command to see what this does. The environment variables
that it sets can be accessed using the $VARIABLE syntax (substuting
VARIABLE for the name of the variable).
$ module show openfoam
Create the top-level user OpenFOAM directory and change to it (not strictly required, but recommended by the tutorials)
$ mkdir -p $FOAM_RUN
$ cd $FOAM_RUN
Tutorial
OpenFOAM comes with several tutorials in the $FOAM_TUTORIALS
directory. This includes the ones in the user
manual.
$ ls $FOAM_TUTORIALS
Until version 11, the standard first tutorial was a cavity flow problem. It is still available in the legacy directory though, and it is one of the simplest examples, so we will use it.
$ cp -r $FOAM_TUTORIALS/legacy/incompressible/icoFoam/cavity/cavity .
$ cd cavity
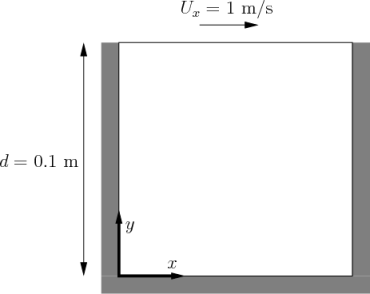
Generate mesh
Generate the mesh data files in constant/polyMesh from the higher-level description file system/blockMeshDict
$ blockMesh
Interactive run
Run the simulation (incompressible flow using the PISO algorithm) in an interactive session to test
$ salloc --mem-per-cpu=500M --ntasks=1 --account=def-SPONSOR
$ icoFoam
$ exit
Batch run
Create a run.sh (name not important) using nano to do the same
$ nano run.sh
#!/bin/bash
#SBATCH --time=00:05:00
#SBATCH --mem-per-cpu=500M
#SBATCH --ntasks=1
#SBATCH --account=def-SPONSOR
#SBATCH --output=icoFoam-%J.log
icoFoam
Submit the batch file to the cluster to run
$ sbatch run.sh
New case
Make a new cavityMPI case based on the cavity case setup
$ cd ..
$ foamCloneCase cavity cavityMPI
$ cd cavityMPI
Run in parallel
Basic four processor system/decomposeParDict decomposition setup copied from damBreak tutorial
$ cp $FOAM_TUTORIALS/incompressibleVoF/damBreakLaminar/damBreak/system/decomposeParDict
Decompose the cavity across the four processors
$ decomposePar
Run the simulation in parallel using four processors
$ salloc --mem-per-cpu=500M --ntasks=4 --account=def-SPONSOR
$ mpirun icoFoam -parallel
$ exit
Collect the output back into a single file
$ reconstructPar
Prep for ParaView
Create a NAME.foam file to give something to open in ParaView
(name doesn’t matter – just identifies it as an OpenFOAM directory)
$ touch cavity.foam
Can also generate VTK input files
$ foamToVtk