Introduction
In this course we are going to get you using the R tidyverse. Tidyverse is a collection of opinionated R packages sharing an underlying design philosophy, grammar, and data structures. They are the results of many years of research, experimentation, and papers by Hadley Wickham and friends, and give the best experience I have ever had doing Data Science across any programming language (including numpy, pandas, and friends from python). The contents of the course is based on, and borrows liberally from, Hadley’s free online book R for Data Science. You are highly encouraged to check it out too if you want to continue with data science.
Outline
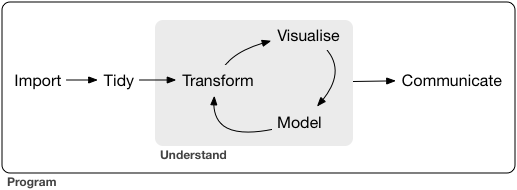
As described in Haley’s book, this first step in data science proojects is importing the data and cleaning it up. The details of what this involve are generally specific the data sources, but typical items would involve dealing with different data formats, missing values, typos and other obvious mistakes that were made during data collection, etc. After beating your data into a format suitable for working with, you can then began an interative process of coming to understand it. Generally this is a repatitive process of visualizing, modeling, and further transforming of the data that gradually aligns your undestanding and the data.
In actualy scientific analysis, it is important to set aside a portion of your data for hypothesis confirmation and not look at this data when coming to your understanding so you have an independent dataset to verify the hypothesis you form. The reason for this is that, while the chance of any one given pattern appearing randomly in a dataset are quite low, the chances of some random pattern appearing in a dataset are quite high. When you are exploring the dataset, you mind looks for any pattern, and, therefore, the probability of it mistaking something random for a pattern is reasonably high. When you verify, however, you are only checking for one specific thing, the probability of one specific thing happening randomly is quite low, and so you can have confidence if it does. You can only do this once though. Sucumming to the temptation to keep verifying hypothesis until one succeeds foils this as you will just really be looking through all your data for any pattern and likely mistaking something random for one. Many published results from big data gene analysis have proven to be incorrect for this reason.
The final step of the process is to come up with a way of using the data to clearly communicate what you have come to understand to others. At a minimmum this will cleaning and expanding on some of the visualizations you produced while studying the data yourself. It may also involve coming up with entirely new visualizations.
Software
In this course, we will be using R. You can either run R directly in a console or (more recommended) the RStudio
environment. You install these on your computer or use the graham VDI machine. R has a large set of add on packages
(including tidyverse). Under Linux, a large selection of these are available pre-build through your computer’s
package manager. We recommend using these if they exist as they are fast to install, installed globally (available
to all user accounts), and less prone to running into issues. Failing that, you can also have R download and build
and package for the local user using the install.packages command.
Personal Windows or Mac OS X
If you want to install it on your computer and are running Windows or Mac OS X, you can download and install
R from the Comprehensive R Archive Network (CRAN) site: https://cran.r-project.org/
RStudio from the from the RStudio site: https://www.rstudio.com/products/rstudio/download/
Once you have installed RStudio, you can start it up and then have R build you the latest tidyverse package from
CRAN (this will take quite awhile)
> install.packages('tidyverse')
Personal Linux
The RCRAN page above also includes instructions on how to install R for the various
common Linux distributions. Most Linux distribution come with a large number of R packages pre-packaged. (i.e.,
installable via apt-get or dnf). You should prefer using these over installing them via the R
install.packages command as they generally have less issues (the system package manager will also install any
other required system dependencies) and get installed globally (instead of just for the current user).
Debian or Ubuntu
For Debian or or Ubuntu you will want to do something like the following (the RStudio link in the following may require updating depending on what is now current and the particular version and number of bits of your Ubuntu Debian installation)
[tyson@tux ~]$ sudo apt-get install r-base r-base-dev r-cran-tidyverse
[tyson@tux ~]$ wget -d https://download1.rstudio.org/desktop/bionic/amd64/rstudio-1.4.1717-amd64.deb
[tyson@tux ~]$ sudo dpkg -i rstudio-1.4.1717-amd64.deb
which installs the system R, the system tidyverse, and a non-system RStudio package downloaded from the RStudio website (Debian and Ubuntu don’t provide RStudio).
Fedora
For Fedora you will want to do something like
[tyson@tux ~]$ sudo dnf install rstudio-desktop
which installs the system R and the system RStudio. Unfortunately Fedora doesn’t currently package the top-level R
tidyverse package, so you have to get R to build and install it
[tyson@tux ~]$ R -e "install.packages('tidyverse',repos='https://utstat.toronto.edu/cran/')"
or be satisifed with a system installation of each of the components (this just means you will have to import each
components separtely instead of altogether with one library(tidyverse) command in R)
[tyson@tux ~]$ sudo dnf install R-ggplot2 R-tibble R-tidyr R-readr R-purrr R-dplyr R-stringr R-forcats
Graham VDI
If you want, you can also use R and RStudio remotely on graham’s virtual desktop interface (VDI) machines. This is especially useful when analyzing data that is already on/being generated on the graham cluster. To access these machines, you need to install and setup the TigerVNC client on your computer as documented on our VNC page on our Compute Canada documentation wiki. A sort summary is that you install the TigerVNC viewer as appropriate for your machine
Windows: install the latest vncviewer executable (exe) from https://sourceforge.net/projects/tigervnc/files/stable
Mac OS X: install the latest TigerVNC dmg from https://sourceforge.net/projects/tigervnc/files/stable
Debian/Ubuntu: run
sudo apt-get install tigervnc-viewerFedora: run
sudo dnf install tigervnc
start it up, enter gra-vdi.computecanada.ca, and pick press the Connect button (if you get a certificate
verification error, see the website for directions on setting up your certificate paths to fix this).
Once logged in with your Compute Canada username and password, you can get a terminal by click the black screen
icon on the bar at the top of the screen. From the terminal you will have access to all your files and the same
software stack as on graham (note that the CcEnv and StdEnv modules are not loaded by default as on
graham).
An easy way to setup R and RStudio environments on gra-vdi is to use the Nix software building and composition
system. Following the R section of the Using Nix page on our
Compute Canada docuumentation wiki, we create an RStudio.nix file in our project directory with a list of the R
packages we want to use (this command does not create the file, it just shows its contents, use an editor like
nano to create it).
[tyson@gra-vdi3 ~]$ cat RStudio.nix
with import <nixpkgs> { };
rstudioWrapper.override {
packages = with rPackages; [
tidyverse
];
}
Then we load the nix module and run the nix run command on the file. This nests a new shell session in our
existing one (type exit to end it) with the PATH environment variable expanded to include the rstudio
wrapper, which enables us to directly launch RStudio.
[tyson@gra-vdi3 ~]$ module load nix
[tyson@gra-vdi3 ~]$ nix run -f RStudio.nix
[tyson@gra-vdi3 ~]$ rstudio
Nix packages most R packages, and, for the same reasons as discussed above, these should be preferred over manually
installing and building packages with the R install.packages command. To change the package set, update the
packages = with rPackages; [ ... ] lines in the RStudio.nix file, exit the existing nix run session, start a
new one with the new package set, and restart RStudio
[tyson@gra-vdi3 ~]$ nano RStudio.nix
[tyson@gra-vdi3 ~]$ exit
[tyson@gra-vdi3 ~]$ nix run -f RStudio.nix
[tyson@gra-vdi3 ~]$ rstudio
As detailed on the Using Nix page, the nix run command only
builds gives a temporary environment guarnateed to last for a day. For longer term environments, use the nix build or the nix-env commands as also documented on the Using Nix page. The former gives a per-project solution
by creating a direct link in your project directory to the R/RStudio wrappers. The later gives a per-user solution
by adding it to your path anytime the Nix module is loaded.
Website
The tidyverse website tidyverse.org contains documentation, examples, and quick
reference cards for each of the tidyverse packages. You are highly encouraged to reference it, especially the quick
reference cards, but be aware that the front page does not show the forcats and stringr package links on
limited screen widths, and the packages menu item has to be used to get to these.